Quinores3D - Quinolone resistance
Antibiotic resistance
Antibiotic resistance is a natural process whereby microorganisms acquire the ability to resist the effects of antimicrobial drugs, due to the selective pressure that results from exposure to these compounds.
Quinolones
Quinolones and fluoroquinolones are a family of broad spectrum, systemic antibacterial agents that have been used widely as therapy of respiratory and urinary tract infections. Fluoroquinolones are active against a wide range of aerobic gram-positive and gram-negative organisms.
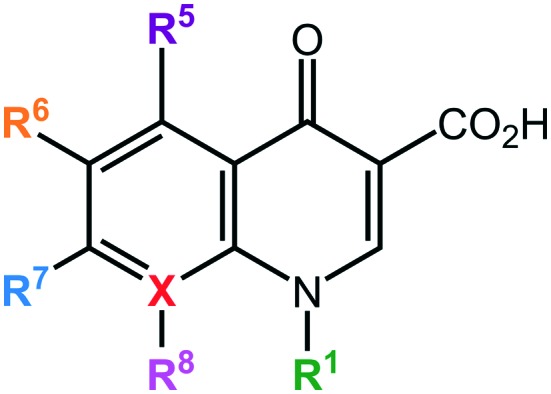
They contain a bicyclic core structure related to the compound 4-quinolone:
The quinolone class of antibiotics inhibits the DNA synthesis of bacteria by targeting two essential bacterial type II topoisomerase enzymes, DNA gyrase (composed of 2 GyrA and 2 GyrB subunits) and DNA topoisomerase IV (composed of 2 ParC and 2 ParE subunits). These two enzymes are critical bacterial enzymes that regulate the chromosomal supercoiling required for DNA synthesis. Both enzymes act via a double-stranded DNA break involving formation of a covalent enzyme–DNA cleavage complex through an ATP-dependent reaction. GyrA corresponds to ParC subunit and they compose the active site, while GyrB corresponds to ParE and they compose the ATPase site.
The drug binds these cleavage complexes at the enzyme–DNA interface in the active site forming a ternary complex of drug, enzyme, and DNA, which leads the DNA replication machinery to block at the replication forks, resulting in an inhibition of DNA synthesis (bacteriostatic effect) and cell death due to lethal double-strand DNA break.
Mechanisms of resistance
Resistance to quinolones can arise due to different mechanisms, in particular point mutations in the quinolone targets, efflux pumps, downregulations of porins in the cell membrane, plasmid carried genes of resistance.
Single amino acid changes in either gyrase or topoisomerase IV can cause quinolone resistance, weakening the interaction between the drugs and enzymes, reducing quinolone susceptibility.These resistance mutations have most commonly been localized to the amino terminal domains of GyrA or ParC and are in proximity to the active site. Mutations in specific domains of GyrB and ParE have also been shown to cause quinolone resistance, although they are substantially less common in resistant clinical bacterial isolates than mutations in GyrA or ParC.